Electrophysiology
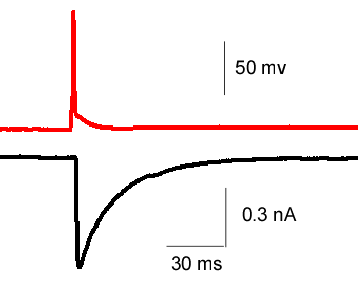
- Postsynaptic current (EPSC) evoked by a presynaptic AP (paired recording)
We employ a variety of electrophysiological techniques to
- characterize the molecular and biophysical properties of membrane proteins, ion channels, transporters and GPCRs (including conformational changes ("gating"), permeation and binding of ions and/or ligands),
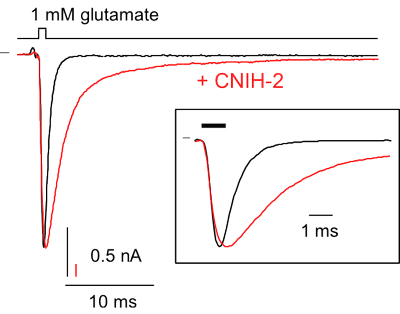
- analyze their interaction with auxiliary subunits and other associated proteins and
- precisely monitor the time-course of protein-protein interactions and their significance for the signal transduction at the plasma membrane.
For these purposes, we use the following techniques and recording configurations:
- patch-clamp recordings in whole-cell, and excised-patch (inside-out, outsice-out) configuration,
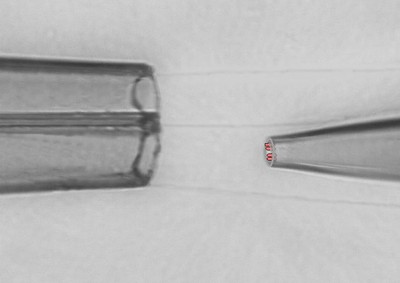
- giant patch-clamp recordings (patch-pipettes with Ø of up to 20 µm)
- two-electrode voltage- and current-clamp,
- rapid piezo-driven application of agonists (ligand-controlled channels and receptors)
- lock-in capacitance measurements (changes in membrane surface associated with exo- and endocytosis).
These techniques are applied on membrane proteins and protein complexes that are
- heterologously expressed in culture-cells (CHO, HEK, COS) and Xenopus oocytes,
- present in native cells, mostly CNS neurons in brain slices and/or neuronal cultures, before and after molecular and or genetic manipulations (eg. virally-driven overexpression of mutant protein, protein knock-down via sh-RNAs).