Membrane Proteomics
 Functional proteomic analysis plays a
central role in modern systems biology and aims at comprehensive
identification and characterization of protein protein interactions,
i. e. the assembly of proteins into complexes and networks as a
general functional and structural organisation principle.
Functional proteomic analysis plays a
central role in modern systems biology and aims at comprehensive
identification and characterization of protein protein interactions,
i. e. the assembly of proteins into complexes and networks as a
general functional and structural organisation principle.
In the projects of our research group functional proteomics is primarily used to unravel and characterize components and mechanisms of fast signal transduction processes at membranes which includes constitutive as well as transient or dynamic (membrane) protein interactions.
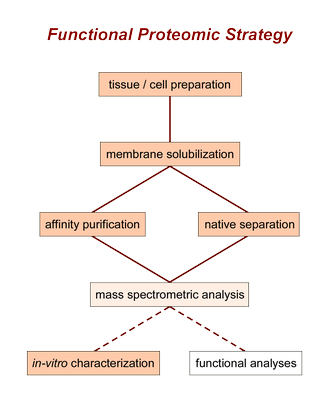
For this purpose we use a number of biochemical techniques which have been systematically developed and standardized in our laboratory as part of a basal strategy:
- solubilization, isolation and separation starting from native tissue
- mass spectrometric identification and quantification
- heterologous reconstitution and biochemical in-vitro characterization
This strategy has been designed to achieve a maximum degree of reliability, sensitivity, closeness to the physiological situation, and rather complete identification of the proteins involved.
Membrane protein solubilization
Biochemical analysis of membrane proteins generally requires the use of detergents to make target proteins and protein complexes soluble. The challenge is to find a reasonable balance between high solubilization efficiency and preservation of structural integrity of proteins and protein assemblies. We monitor these parameters using different assays (Western blot analysis, native gel electrophoresis, binding assays) with a range of buffer systems to finally determine the optimal conditions. In doing so we benefit from the know-how and ComplexioLyte buffers of our collaboration partner Logopharm.
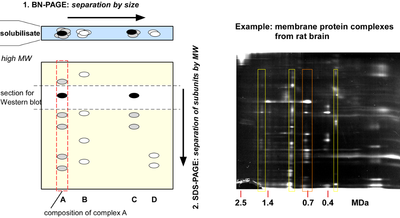
Native gel electrophoresis
Separation of protein complexes by native gel electrophoresis, originally developed by Schaegger and coworkers as blue native PAGE (BNPage) for analysis of mitochondrial membrane protein complexes, is one of the few options for high resolution separation of large protein complexes. Separation is achieved electrophoretically in two dimensions: by molecular (complex) size (0.1-10 MDa) along a pore gradient gel under native conditions and, after successive denaturation of the complexes, in a second dimension by SDS-PAGE according to the molecular weight of the corresponding subunits. The size distribution of target complexes as visualized by Western blot or mass spectrometric analysis, which may also be manipulated by addition of target-specific antibodies (so-called shift assays) allows for qualitative as well as quantitative conclusions about composition and stability of protein complexes.
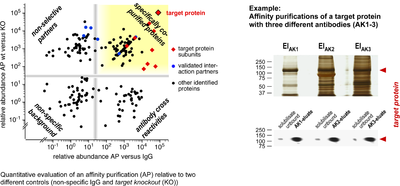
Affinity purification
- Quantitative evaluation of an affinity purification and example of a target protein with different antibodies
Target proteins are isolated together with their associated partners by affinity capture, namely by using specific antibodies. Despite their generally high specificity and affinity, antibodies also have a number of adverse properties that can lead to significant errors in interaction analyses. Therefore, we use different stringent controls, such as target knockout tissue, and compare the results from multiple antibodies directed against the same target protein (multi epitope approach). When combined with quantitative mass spectrometric analysis, also rare or dynamic interaction partners can be identified with low error rates.
For a more detailed biochemical in-vitro characterization of protein-protein interactions we use a number of specialized techniques in addition: Protein complexes are reconstituted by co-expression in heterologous systems, in-vitro co-translation or in binding assays. Stability and stoichiometry of protein-protein interactions are analyzed by antibody shift assays, co-sedimentation centrifugation, analytical ultracentrifugation, gel filtration, photometric binding assays and quantitative mass spectrometry.


 Previous:
Electrophysiology
Previous:
Electrophysiology
